[1]:
import os, sys
import numpy as np
import corner
# sys.path.append('../')
import py21cmfish as p21fish
import matplotlib as mpl
import matplotlib.pyplot as plt
import matplotlib.mathtext as mathtext
import matplotlib.lines as mlines
%matplotlib inline
plt.style.use(['default','seaborn','seaborn-ticks'])
mpl.rcParams['xtick.direction'] = 'in'
mpl.rcParams['ytick.direction'] = 'in'
mpl.rcParams['figure.figsize'] = (4,3)
mpl.rcParams['figure.dpi'] = 150
if os.path.exists(os.environ['WORK_DIR']+'/code/matplotlibrc'):
from matplotlib import rc_file
rc_file(os.environ['WORK_DIR']+'/code/matplotlibrc')
mathtext.FontConstantsBase.sup1 = 0.5
mathtext.FontConstantsBase.sub1 = 0.2
mathtext.FontConstantsBase.sub2 = 0.2
[3]:
# Color palette
try:
from palettable.tableau import Tableau_20, ColorBlind_10
cols = ColorBlind_10.hex_colors
col_pess = cols[6]
col_mod = cols[0]
col_alpha = 'k'
col_mcmc = cols[3]
col_P19 = cols[1]
except:
col_pess = '0.5'
col_mod = 'tab:blue'
col_alpha = 'k'
col_mcmc = '0.7'
col_P19 = 'tab:orange'
[4]:
%load_ext autoreload
%autoreload 2
Fisher plots
This notebook loads and plots posteriors based on the 21cm power spectrum for the 21cmfish paper.
To run the notebook you must first unpack the data directories in /examples/
[23]:
p21fish.base_path = '/Users/cmason/Documents/Research/21cmFAST/21cmfish/'
examples_dir = p21fish.base_path+'examples/'
data_dir = examples_dir+'data/'
noise_dir = data_dir+'21cmSense_noise/'
figs_dir = examples_dir
print('Loading data from',data_dir)
Loading data from /Users/cmason/Documents/Research/21cmFAST/21cmfish/examples/data/
EOS21
This is a fiducial case from Munoz+2021 with CDM, and with both pop II and pop III galaxies.
[7]:
# Find the parameters we varied and fiducials from the config file
# but you could also list these yourself (especially if you want to change the order)
print(p21fish.base_path+'21cmFAST_config_files/EoS_mini.config')
astro_params_vary, astro_params_fid = p21fish.get_params_fid(
config_file=p21fish.base_path+'21cmFAST_config_files/EoS_mini.config')
print('Varying parameters:',astro_params_vary)
print('Fiducial parameter values:',astro_params_fid)
assert type(astro_params_vary) == list, 'astro_params_vary must be a list'
assert type(astro_params_fid) == dict, 'astro_params_vary must be a dict'
/Users/cmason/Documents/Research/21cmFAST/21cmfish/21cmFAST_config_files/EoS_mini.config
Varying parameters: ['ALPHA_STAR', 'F_STAR10', 'ALPHA_ESC', 'F_ESC10', 'ALPHA_STAR_MINI', 'F_STAR7_MINI', 'F_ESC7_MINI', 'L_X', 'NU_X_THRESH', 'A_LW']
Fiducial parameter values: {'ALPHA_ESC': -0.3, 'F_ESC10': -1.35, 'ALPHA_STAR': 0.5, 'F_STAR10': -1.25, 't_STAR': 0.5, 'F_STAR7_MINI': -2.5, 'ALPHA_STAR_MINI': 0.0, 'F_ESC7_MINI': -1.35, 'L_X': 40.5, 'L_X_MINI': 40.5, 'NU_X_THRESH': 500.0, 'A_VCB': 1.0, 'A_LW': 2.0}
Load parameters
Moderate noise
[9]:
# Load each parameter into a dictionary
params_EoS = {}
astro_params_vary_EoS = ['F_STAR10', 'ALPHA_STAR', 'F_ESC10', 'ALPHA_ESC',
'F_STAR7_MINI', 'ALPHA_STAR_MINI', 'F_ESC7_MINI', 'A_LW', 'L_X', 'NU_X_THRESH',]
for param in astro_params_vary_EoS:
params_EoS[param] = p21fish.Parameter(param=param,
output_dir=data_dir+'EOS21/',
PS_err_dir=noise_dir+'21cmSense_fid_EOS21/',
clobber=False, Park19=None,
vb=False, new=False)
########### fisher set up for F_STAR10
Loading global signal and power spectra from saved files
Fiducial: F_STAR10=-1.25
########### fisher set up for ALPHA_STAR
Loading global signal and power spectra from saved files
Fiducial: ALPHA_STAR=0.5
########### fisher set up for F_ESC10
Loading global signal and power spectra from saved files
Fiducial: F_ESC10=-1.35
########### fisher set up for ALPHA_ESC
Loading global signal and power spectra from saved files
Fiducial: ALPHA_ESC=-0.3
########### fisher set up for F_STAR7_MINI
Loading global signal and power spectra from saved files
Fiducial: F_STAR7_MINI=-2.5
########### fisher set up for ALPHA_STAR_MINI
Loading global signal and power spectra from saved files
Fiducial: ALPHA_STAR_MINI=0.0
########### fisher set up for F_ESC7_MINI
Loading global signal and power spectra from saved files
Fiducial: F_ESC7_MINI=-1.35
########### fisher set up for A_LW
Loading global signal and power spectra from saved files
Fiducial: A_LW=2.0
########### fisher set up for L_X
Loading global signal and power spectra from saved files
Fiducial: L_X=40.5
########### fisher set up for NU_X_THRESH
Loading global signal and power spectra from saved files
Fiducial: NU_X_THRESH=500.0
Pessimistic noise
[10]:
# Load each parameter into a dictionary
params_EoS_pess = {}
for param in astro_params_vary_EoS:
params_EoS_pess[param] = p21fish.Parameter(param=param,
output_dir=data_dir+'EOS21/',
PS_err_dir=noise_dir+'21cmSense_pess_EOS21/',
clobber=False, Park19=None,
vb=False)
########### fisher set up for F_STAR10
Loading global signal and power spectra from saved files
Fiducial: F_STAR10=-1.25
########### fisher set up for ALPHA_STAR
Loading global signal and power spectra from saved files
Fiducial: ALPHA_STAR=0.5
########### fisher set up for F_ESC10
Loading global signal and power spectra from saved files
Fiducial: F_ESC10=-1.35
########### fisher set up for ALPHA_ESC
Loading global signal and power spectra from saved files
Fiducial: ALPHA_ESC=-0.3
########### fisher set up for F_STAR7_MINI
Loading global signal and power spectra from saved files
Fiducial: F_STAR7_MINI=-2.5
########### fisher set up for ALPHA_STAR_MINI
Loading global signal and power spectra from saved files
Fiducial: ALPHA_STAR_MINI=0.0
########### fisher set up for F_ESC7_MINI
Loading global signal and power spectra from saved files
Fiducial: F_ESC7_MINI=-1.35
########### fisher set up for A_LW
Loading global signal and power spectra from saved files
Fiducial: A_LW=2.0
########### fisher set up for L_X
Loading global signal and power spectra from saved files
Fiducial: L_X=40.5
########### fisher set up for NU_X_THRESH
Loading global signal and power spectra from saved files
Fiducial: NU_X_THRESH=500.0
Fisher matrix analysis
make_fisher_matrix() creates the Fisher matrix and its inverse from a Parameters dictionary. The resulting ellipses can be plotted with plot_triangle().
[11]:
Fij_matrix_PS, Finv_PS = p21fish.make_fisher_matrix(params_EoS, fisher_params=astro_params_vary_EoS,
hpeak=0.0, obs='PS',
k_min=0.1, k_max=1,
z_min=5., z_max=30.,
sigma_mod_frac=0.2,
add_sigma_poisson=True)
fid_params = np.array([astro_params_fid[param] for param in params_EoS])
fid_labels = np.array([p21fish.astro_params_labels[param] for param in params_EoS])
PS shape: (23, 24)
[12]:
p21fish.plot_triangle(params=astro_params_vary_EoS,
fiducial=fid_params,
labels=fid_labels,
cov=Finv_PS,
ellipse_color=col_mod,
title_fontsize=14,
xlabel_kwargs={'labelpad': 5, 'fontsize':22},
ylabel_kwargs={'labelpad': 5, 'fontsize':22},
fig_kwargs={'figsize':(18,18)});
plt.savefig(figs_dir+'corner_EoS_mini_fisher.png', bbox_inches='tight')
generating new axis
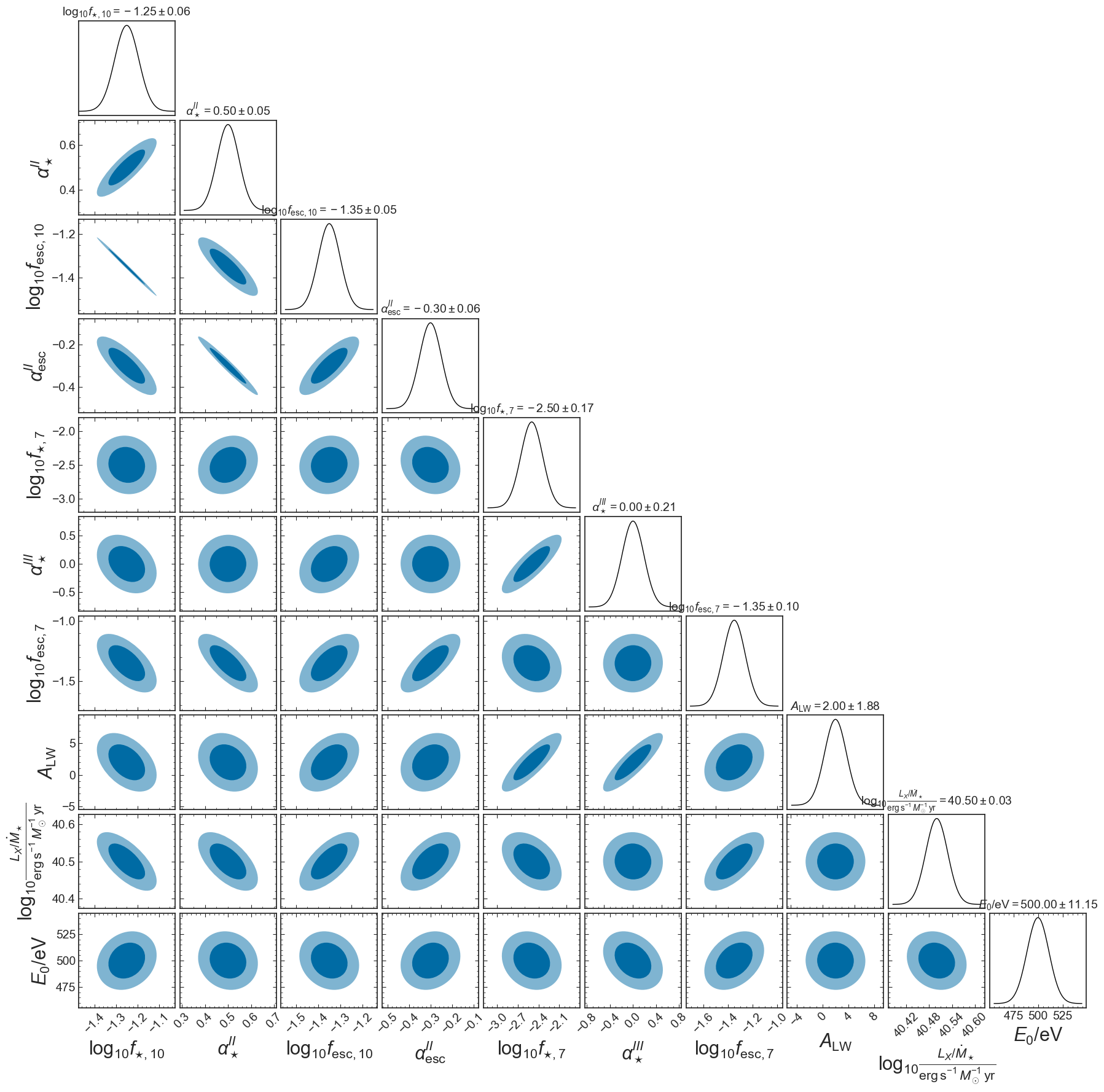
Add a prior
E.g. Park+2019 find \(\sigma(\alpha_\star^{II}) \approx 0.07\).
To add a prior, we can add 1/\(\sigma^2\) to the diagonal element for that parameter (e.g. Coe 2009)
[13]:
sigma_alpha_star_II = 0.07
idx_alpha_star = list(params_EoS).index("ALPHA_STAR")
print(f'ALPHA_STAR is at index={idx_alpha_star}')
Fij_matrix_PS_alpha_star_prior = Fij_matrix_PS.copy()
Fij_matrix_PS_alpha_star_prior[idx_alpha_star,idx_alpha_star] += 1/sigma_alpha_star_II**2.
Finv_alpha_star_prior = np.linalg.inv(Fij_matrix_PS_alpha_star_prior)
ALPHA_STAR is at index=1
[14]:
fig, ax = plt.subplots(len(fid_params), len(fid_params), figsize=(18,18))
cols = [col_mod, col_alpha]
for i, cov in enumerate([Finv_PS, Finv_alpha_star_prior]):
col = cols[i]
if i == 0:
# No prior
resize_lims=True
ellipse_color=col
ellipse_kwargs=[{'alpha':0.8},{'alpha':0.2}]
plot1D_kwargs={'c':col, 'lw':1}
else:
# with prior
ls='solid'
resize_lims=False
ellipse_color='None'
ellipse_kwargs=[{'edgecolor':col,'lw':2,'ls':ls},
{'edgecolor':col,'lw':2,'ls':ls,'alpha':0.8}]
plot1D_kwargs={'c':col, 'lw':2, 'ls':ls}
p21fish.plot_triangle(params=astro_params_vary_EoS,
fiducial=fid_params,
labels=fid_labels,
cov=cov,
ellipse_color=ellipse_color,
ellipse_kwargs=ellipse_kwargs,
plot1D_kwargs=plot1D_kwargs,
resize_lims=resize_lims,
title_fontsize=14,
xlabel_kwargs={'labelpad': 5, 'fontsize':22},
ylabel_kwargs={'labelpad': 5, 'fontsize':22},
ax=ax, fig=fig);
p21fish.title_double_ellipses(axes=ax, labels=fid_labels,
med=fid_params, sigma=np.sqrt(cov.diagonal()),
title_fontsize=18, title_pad=55,
vspace=i/5,
color=col
)
no_prior = mlines.Line2D([], [], color=col_mod, lw=2, label='EOS, 21cm only')
w_prior = mlines.Line2D([], [], color=col_alpha, lw=3, ls=ls, label=r'+ LF $\alpha_\star^\mathrm{II}$ prior')
ax[0,5].legend(handles=[no_prior, w_prior], loc='upper left', fontsize=24)
plt.savefig(figs_dir+'corner_EoS_mini_fisher_ALPHA_STAR_prior.png', bbox_inches='tight')
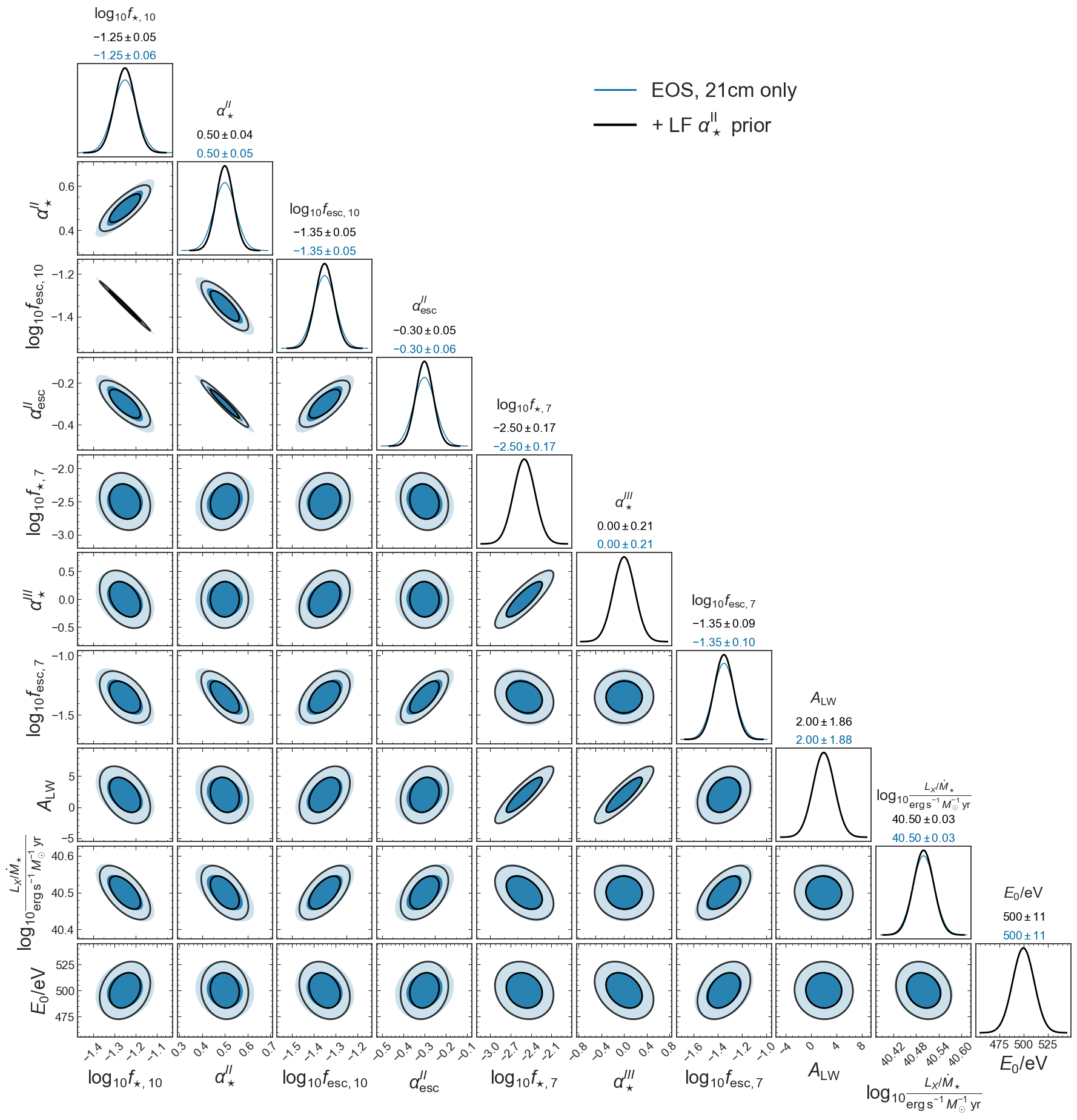
Pessimistic noise case
[15]:
Fij_matrix_PS_pess, Finv_PS_pess = p21fish.make_fisher_matrix(params_EoS_pess,
fisher_params=astro_params_vary_EoS,
hpeak=0.0, obs='PS',
k_min=0.1, k_max=1,
z_min=5., z_max=30.,
sigma_mod_frac=0.2,
add_sigma_poisson=True)
PS shape: (23, 24)
[16]:
fig, ax = plt.subplots(len(fid_params), len(fid_params), figsize=(18,18))
cols = [col_pess, col_mod]
for i, cov in enumerate([Finv_PS_pess, Finv_PS]):
col = cols[i]
if i == 0:
# Pessimistic
resize_lims=True
plot_rescale=2
ellipse_color='None'
ellipse_kwargs=[{'edgecolor':col,'lw':1,'ls':'dashed'},
{'edgecolor':col,'lw':1,'ls':'dashed','alpha':1}]
plot1D_kwargs={'c':col, 'lw':1, 'ls':'dashed'}
else:
# Moderate
ls='solid'
resize_lims=False
plot_rescale=4
ellipse_color=col
ellipse_kwargs=[{},{'alpha':0.5}]
plot1D_kwargs={'c':col, 'lw':2}
p21fish.plot_triangle(params=astro_params_vary_EoS,
fiducial=fid_params,
labels=fid_labels,
cov=cov,
ellipse_color=ellipse_color,
ellipse_kwargs=ellipse_kwargs,
plot1D_kwargs=plot1D_kwargs,
resize_lims=resize_lims,
plot_rescale=plot_rescale,
title_fontsize=14,
xlabel_kwargs={'labelpad': 5, 'fontsize':22},
ylabel_kwargs={'labelpad': 5, 'fontsize':22},
ax=ax, fig=fig);
p21fish.title_double_ellipses(axes=ax, labels=fid_labels,
med=fid_params, sigma=np.sqrt(cov.diagonal()),
title_fontsize=18, title_pad=50,
vspace=i/5, color=col)
no_prior = mlines.Line2D([], [], color=col_pess, lw=2, ls='dashed', label='Pessimistic foregrounds')
w_prior = mlines.Line2D([], [], color=col_mod, lw=4, label=r'Moderate foregrounds')
ax[0,5].legend(handles=[w_prior, no_prior], loc='upper left', fontsize=28)
plt.savefig(figs_dir+'corner_EoS_mini_fisher_pessimistic.png', bbox_inches='tight')
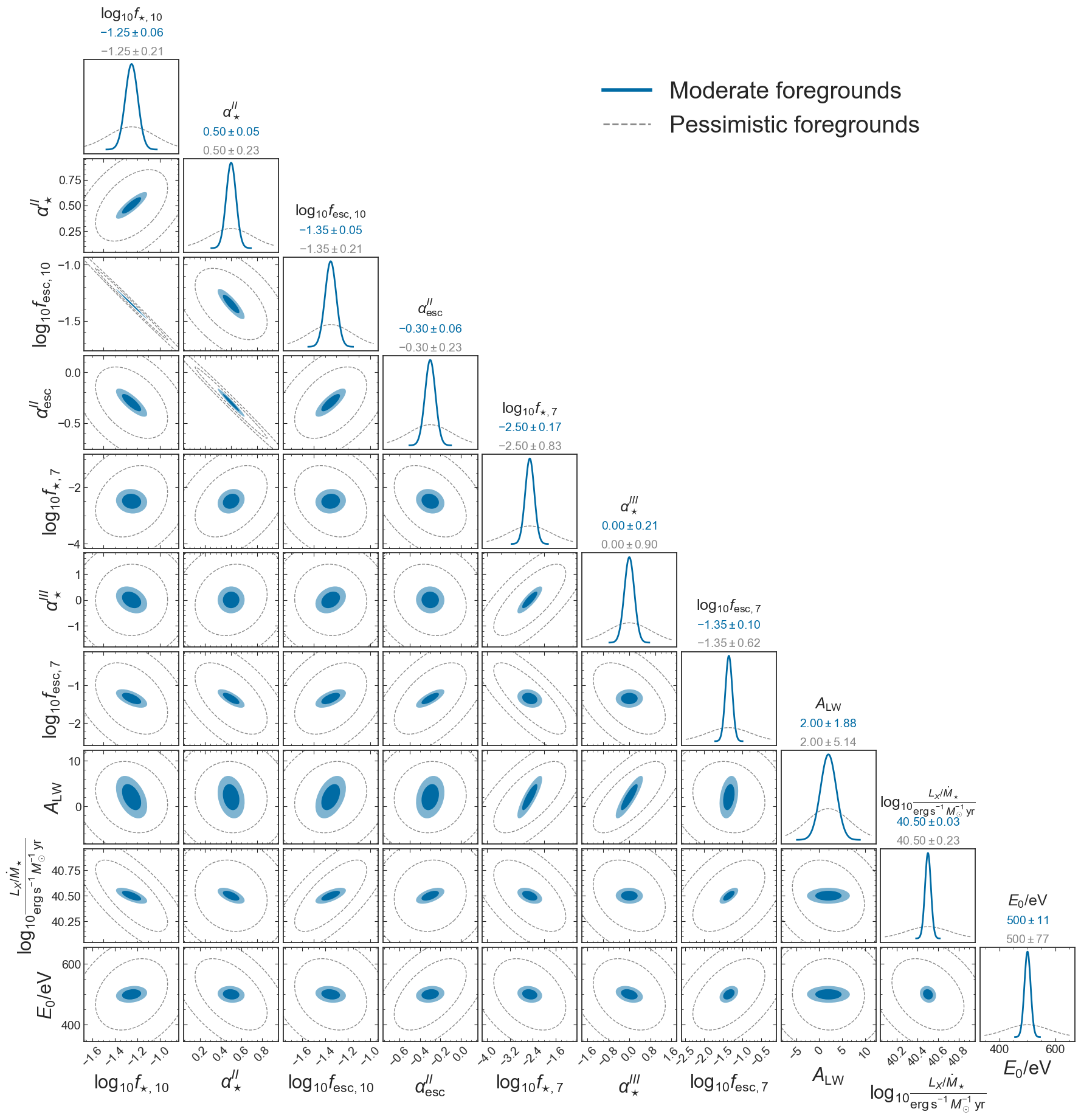
Comparison of forecasts
S/N
[17]:
param_test = params_EoS['ALPHA_ESC']
param_test_pess = params_EoS_pess['ALPHA_ESC']
SNR = np.zeros((len(param_test.PS_z_HERA),2))
for i in range(len(param_test.PS_z_HERA)):
PS = param_test.PS['CDM']['ALPHA_ESC=-0.3'][i]['delta']
k = param_test.PS['CDM']['ALPHA_ESC=-0.3'][i]['k'][~np.isnan(PS)]
PS = PS[~np.isnan(PS)]
# Get rid of nans and interp PS on HERA grid
PS_interp = np.interp(param_test.PS_err[i]['k']*0.7,
k, PS)
# Use HERA errors
PS_err = np.array([param_test.PS_err[i]['err_mod'],
param_test_pess.PS_err[i]['err_mod']])
SNR[i] = np.sqrt(np.sum((PS_interp/PS_err)**2., axis=1))
SNR_EoR = np.sqrt(np.sum(SNR[:,0][param_test.PS_z_HERA<7.]**2.)) # ~300
SNR_CD = np.sqrt(np.sum(SNR[:,0][param_test.PS_z_HERA>=7.]**2.)) # ~300
print(SNR_EoR, SNR_CD)
plt.figure(figsize=(6.5,4))
labels = ['Moderate','Pessimistic']
cols = [col_mod, col_pess]
lss = ['solid', 'dashed']
for i, s in enumerate(SNR.T):
SNR_total = np.sqrt(np.sum(s**2.)) # ~300
plt.plot(param_test.PS_z_HERA, s, c=cols[i], ls=lss[i], label=f'{labels[i]} foregrounds, total S/N = {SNR_total:.0f}')
plt.xlabel('Redshift, z')
plt.ylabel('HERA $\Delta_{21}^2$ S/N')
plt.annotate("Reionization", xy=(7, 20), xycoords='data',
xytext=(7, 0.6), ha='center', fontsize=10,
arrowprops=dict(arrowstyle="->", lw=0.5))
plt.annotate("Epoch of\nHeating", xy=(12, 4), xycoords='data',
xytext=(12, 0.1), ha='center', fontsize=10,
arrowprops=dict(arrowstyle="->", lw=0.5))
plt.annotate("Cosmic\nDawn", xy=(18, 0.5), xycoords='data',
xytext=(18, 0.05), ha='center', fontsize=10,
arrowprops=dict(arrowstyle="->", lw=0.5))
plt.axhline(10., lw=0.5, ls='solid', c='0.5', zorder=0)
plt.legend()
plt.yscale('log')
plt.savefig(figs_dir+'SNR_EoS.pdf', bbox_inches='tight')
218.50718001785071 285.78849803538145
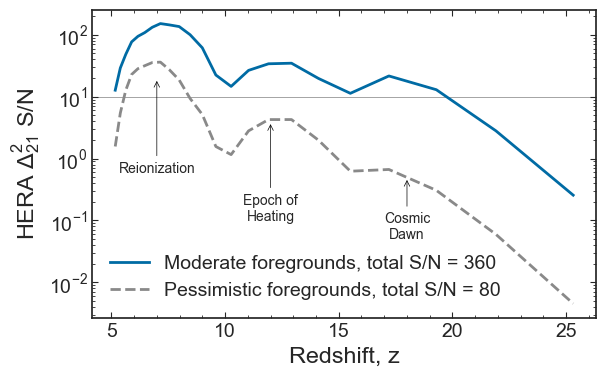
Errors for each parameter
[18]:
cols = [col_mod, col_pess, col_mod, col_pess]
lss = ['solid', 'dashed','solid', 'dashed']
lws = [3,3,1.5,1.5]
markers = ['o','s','o','s']
labels = ['Moderate foregrounds','Pessimistic foregrounds',r'+ LF $\alpha_\star^\mathrm{II}$ prior','']#r'with LF $\alpha_\star^\mathrm{II}$ prior']
alphas = [1,1,0.6,0.6]
# Add alpha star II prior
sigma_alpha_star_II = 0.07
idx_alpha_star = list(params_EoS).index("ALPHA_STAR")
print(f'ALPHA_STAR is at index={idx_alpha_star}')
Fij_matrix_PS_alpha_star_prior_pess = Fij_matrix_PS_pess.copy()
Fij_matrix_PS_alpha_star_prior_pess[idx_alpha_star,idx_alpha_star] += 1/sigma_alpha_star_II**2.
Finv_alpha_star_prior_pess = np.linalg.inv(Fij_matrix_PS_alpha_star_prior_pess)
# Fractional error
plt.figure(figsize=(7,4))
for cc, cov in enumerate([Finv_PS, Finv_PS_pess, Finv_alpha_star_prior, Finv_alpha_star_prior_pess]):
mean = fid_params
err = np.sqrt(np.diag(cov))
frac_err = np.abs(err/mean)
frac_err[np.isinf(frac_err)] = err[np.isinf(frac_err)]#/0.01
plt.semilogy(fid_labels, frac_err, c=cols[cc], marker=markers[cc],
lw=lws[cc], ls=lss[cc], label=labels[cc], ms=7, alpha=alphas[cc])
# plt.ylim(0.,2.)
plt.axhline(0.1, lw=1, c='k', zorder=0)
plt.legend()
plt.gca().set_xticklabels(fid_labels, rotation = 90, ha="center")
plt.minorticks_on()
plt.gca().tick_params(axis='x', which='minor', bottom=False, top=False)
plt.ylabel('Fractional error')
plt.ylabel(r'$|\sigma(\theta)/\theta_\mathrm{fid}|$')
plt.savefig(figs_dir+'fraction_error_EoS.pdf', bbox_inches='tight')
ALPHA_STAR is at index=1
/var/folders/sy/j00h6jjd19z46r9qd031n3080000gn/T/ipykernel_46372/693560164.py:22: RuntimeWarning: divide by zero encountered in true_divide
frac_err = np.abs(err/mean)
/var/folders/sy/j00h6jjd19z46r9qd031n3080000gn/T/ipykernel_46372/693560164.py:22: RuntimeWarning: divide by zero encountered in true_divide
frac_err = np.abs(err/mean)
/var/folders/sy/j00h6jjd19z46r9qd031n3080000gn/T/ipykernel_46372/693560164.py:22: RuntimeWarning: divide by zero encountered in true_divide
frac_err = np.abs(err/mean)
/var/folders/sy/j00h6jjd19z46r9qd031n3080000gn/T/ipykernel_46372/693560164.py:22: RuntimeWarning: divide by zero encountered in true_divide
frac_err = np.abs(err/mean)
/var/folders/sy/j00h6jjd19z46r9qd031n3080000gn/T/ipykernel_46372/693560164.py:31: UserWarning: FixedFormatter should only be used together with FixedLocator
plt.gca().set_xticklabels(fid_labels, rotation = 90, ha="center")
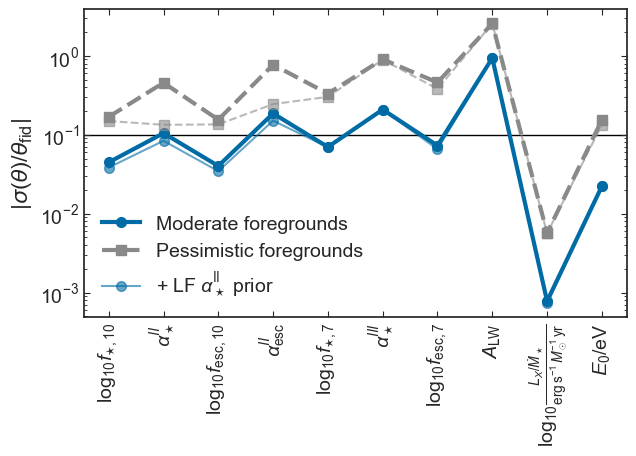
Parameter uncertainty as a function of z
How does noise on each parameter change as a function of z?
[20]:
# Rolling loop through HERA z_bins
z_bins = params_EoS['ALPHA_STAR'].PS_z_HERA
bin_i = 2
z_err = np.zeros((len(z_bins)-bin_i,len(params_EoS)))
for zz in range(len(z_bins)):
if zz < len(z_bins) - bin_i:
Fij_matrix_PS_z, Finv_PS_z = p21fish.make_fisher_matrix(params_EoS,
fisher_params=astro_params_vary_EoS,
hpeak=0.0, obs='PS',
k_min=0.1, k_max=1,
z_min=z_bins[zz], z_max=z_bins[zz+bin_i],
sigma_mod_frac=0.2,
add_sigma_poisson=True)
z_err[zz] = np.sqrt(np.diag(Finv_PS_z))
try:
print(z_err[zz])
except:
print(f'could not do for z_min={z_bins[zz]}')
PS shape: (3, 24)
[1.73627339e-01 3.21997284e-01 1.64274933e-01 3.54371796e-01
6.43228227e-01 2.18440916e+00 9.14333462e-01 5.47960990e+00
3.31960353e+00 1.41965402e+03]
PS shape: (3, 24)
[5.69850700e-01 4.33426673e-01 5.74504689e-01 4.71365660e-01
9.38422255e-01 2.81596463e+00 1.19019524e+00 5.66340515e+00
3.30127823e+00 1.41117918e+03]
PS shape: (3, 24)
[9.57823876e-01 6.25345235e-01 9.83876034e-01 6.19645407e-01
2.09700055e+00 3.04682008e+00 2.35507809e+00 9.08661934e+00
4.95058623e+00 2.28170497e+03]
PS shape: (3, 24)
[1.29039754e+00 1.11743289e+00 1.28640278e+00 1.09660340e+00
2.43973823e+00 3.56788641e+00 2.20000473e+00 1.01747053e+01
5.53384048e+00 3.14006363e+03]
PS shape: (3, 24)
[1.42347618e+00 1.02975807e+00 1.42237059e+00 1.00031553e+00
2.35255424e+00 3.45803485e+00 2.42988755e+00 1.01283615e+01
3.89155274e+00 2.44288794e+03]
PS shape: (3, 24)
[2.00898199e+00 1.03708211e+00 2.05638690e+00 1.00663800e+00
2.80485967e+00 3.93572488e+00 2.85483480e+00 1.14190180e+01
4.02622973e+00 2.94352950e+03]
PS shape: (3, 24)
[2.83314797e+00 1.04695408e+00 2.92667762e+00 1.07212043e+00
3.37826162e+00 4.55540694e+00 3.16791512e+00 1.18427086e+01
4.41766419e+00 3.82393954e+03]
PS shape: (3, 24)
[2.83715369e+00 1.27069997e+00 2.93067068e+00 1.29798668e+00
3.37881859e+00 4.15470244e+00 3.35996461e+00 1.13491570e+01
2.99187043e+00 1.62406559e+03]
PS shape: (3, 24)
[2.42690839e+00 9.69404819e-01 2.49015381e+00 1.00789548e+00
3.22585525e+00 3.29751587e+00 3.35672250e+00 1.04717535e+01
2.55887591e+00 1.15416093e+03]
PS shape: (3, 24)
[ 2.18895132 0.89452448 2.26287453 0.82882134 2.89809027
3.00959964 2.6665641 7.97997123 2.14463165 511.11476398]
PS shape: (3, 24)
[ 2.30253173 1.09834841 2.39621442 0.7044284 2.7189927
4.54343258 2.35622739 6.57932659 2.0492352 181.71146417]
PS shape: (3, 24)
[ 2.47398262 1.38578634 2.51251628 0.77094446 3.27952769
5.495576 2.45835311 7.7119268 1.9562515 197.2576004 ]
PS shape: (3, 24)
[ 2.10965978 1.66444569 2.14777429 1.04804706 3.15203387
5.87810564 2.36037784 9.31892967 1.44931999 153.08614851]
PS shape: (3, 24)
[ 3.45878663 2.63031743 3.90317019 2.06343023 4.64134308 7.42270772
3.78534975 23.18942052 2.13982131 84.52774824]
PS shape: (3, 24)
[ 4.91226697 3.96552484 4.34806075 3.29412742 6.69401367
6.90566649 5.92364083 51.72607217 3.02892226 166.24944922]
PS shape: (3, 24)
[ 3.33283762 2.69103934 4.19583621 3.80850855 4.99314859
4.65599166 4.08427946 47.87914369 1.49643627 155.18310369]
PS shape: (3, 24)
[ 3.94727591 3.01077852 5.63330895 4.91043253 4.80404203
4.56218837 4.98341338 55.7363171 0.566741 170.11927535]
PS shape: (3, 24)
[ 4.39386608 3.29057927 6.07370211 6.35074796 3.59518085
3.48371375 4.7130184 51.96322023 0.58270932 243.68248413]
PS shape: (3, 24)
[ 7.58682717 5.22683021 7.82772596 5.82829266 3.18337457
3.28843803 4.39499209 56.45200407 1.22915709 583.2666204 ]
PS shape: (3, 24)
[ 37.35097905 21.91121519 24.35661314 12.52172589 6.06648355
5.60374057 9.78753992 106.96693703 2.8663244 1048.04966578]
PS shape: (3, 24)
[ 221.07024477 114.79571427 81.527433 40.03929817 16.86206265
17.62594181 37.04349478 239.18451863 28.11076634 5748.68785231]
could not do for z_min=21.909972214717644
could not do for z_min=25.308702138693505
[22]:
from palettable.colorbrewer.sequential import Blues_5, YlGn_5, RdPu_6
z_plot = (z_bins[bin_i:]+z_bins[bin_i-1:-1])/2
fig1, ax1 = plt.subplots(1,1, figsize=(6,4))
sum_err = np.zeros(len(astro_params_vary_EoS))
for pp,p in enumerate(astro_params_vary_EoS):
if pp < 4:
# popII
ls = 'dashed'
lw = 2
colors = RdPu_6.hex_colors[::-1]
i = pp
alpha=1
elif 3 < pp <= 7:
ls = 'solid'
lw = 3
colors = YlGn_5.hex_colors[::-1]
alpha=0.8
i = pp-4
else:
ls = 'dotted'
lw = 2
colors = Blues_5.hex_colors[::-1]
i = (pp-8)*2
alpha=1
# print(p, pp, i)
frac_err = z_err[:,pp]/fid_params[pp]
frac_err[np.isinf(frac_err)] = z_err[:,pp][np.isinf(frac_err)]
sum_err[pp] = np.sqrt(np.mean(np.abs(frac_err)**2.))
# print(p, sum_err[pp])
ax1.semilogy(z_plot, np.abs(frac_err), label=fid_labels[pp],
c=colors[i], ls=ls, lw=lw, alpha=alpha)
ax1.legend(ncol=3, fontsize=10, handlelength=1.5, handletextpad=0.5, columnspacing=1., loc='upper left')
ax1.set_xlabel('Redshift, z')
ax1.set_ylabel('$\sigma$')
ax1.set_ylabel(r'$|\sigma(\theta, z)/\theta_\mathrm{fid}|$')
plt.plot(z_bins, [0.01]*len(z_bins), '|', color='k')
plt.ylim(7e-3, 2e3)
plt.savefig(figs_dir+'fraction_error_EoS_z.pdf', bbox_inches='tight')
/var/folders/sy/j00h6jjd19z46r9qd031n3080000gn/T/ipykernel_46372/1430737097.py:30: RuntimeWarning: divide by zero encountered in true_divide
frac_err = z_err[:,pp]/fid_params[pp]
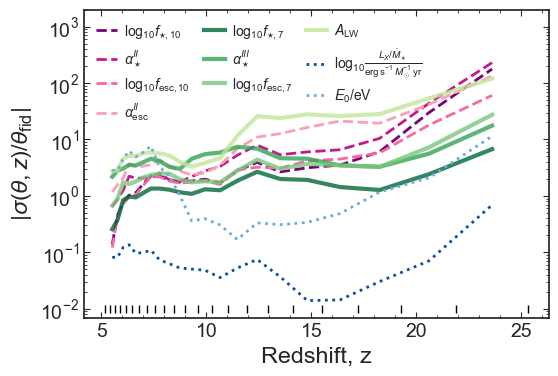
Comparison to Park+19
Compare Fisher matrix with Park+2019 fiducial to their MCMC (21cm power spectrum only)
[24]:
output_dir_Park19 = data_dir+'Park19/'
PS_err_dir_Park19 = noise_dir+'21cmSense_noise_Park19/'
astro_params_vary_Park19, astro_params_fid_Park19 = p21fish.get_params_fid(
config_file=p21fish.base_path+'21cmFAST_config_files/Park19.config')
# Reorder to match Park+19
astro_params_vary_Park19 = ['F_STAR10', 'ALPHA_STAR',
'F_ESC10', 'ALPHA_ESC',
'M_TURN', 't_STAR',
'L_X', 'NU_X_THRESH']
[25]:
# Load parameters
params_Park19 = {}
for param in astro_params_vary_Park19:
params_Park19[param] = p21fish.Parameter(param=param,
output_dir=output_dir_Park19,
HII_DIM=128, BOX_LEN=250,
min_redshift=5.9,
PS_err_dir=PS_err_dir_Park19,
clobber=False, Park19='real',
vb=False, new=False)
########### fisher set up for F_STAR10
Loading global signal and power spectra from saved files
Fiducial: F_STAR10=-1.3
########### fisher set up for ALPHA_STAR
Loading global signal and power spectra from saved files
Fiducial: ALPHA_STAR=0.5
########### fisher set up for F_ESC10
Loading global signal and power spectra from saved files
Fiducial: F_ESC10=-1.0
########### fisher set up for ALPHA_ESC
Loading global signal and power spectra from saved files
Fiducial: ALPHA_ESC=-0.5
########### fisher set up for M_TURN
Loading global signal and power spectra from saved files
Fiducial: M_TURN=8.7
########### fisher set up for t_STAR
Loading global signal and power spectra from saved files
Fiducial: t_STAR=0.5
########### fisher set up for L_X
Loading global signal and power spectra from saved files
Fiducial: L_X=40.5
########### fisher set up for NU_X_THRESH
Loading global signal and power spectra from saved files
Fiducial: NU_X_THRESH=500.0
Make Fisher matrix
[26]:
Fij_matrix_PS_Park19, Finv_PS_Park19 = p21fish.make_fisher_matrix(params_Park19,
fisher_params=astro_params_vary_Park19,
hpeak=0.0, obs='PS',
k_min=0.1, k_max=1,
z_min=5.7, z_max=30.,
sigma_mod_frac=0.2,
cosmo_key='CDM',
add_sigma_poisson=True)
fid_params_Park19 = np.array([astro_params_fid_Park19[param] for param in params_Park19])
fid_labels_Park19 = np.array([p21fish.astro_params_labels[param] for param in params_Park19])
PS shape: (12, 23)
Load Park19 chains and compare
Load their 21cm-only chains and compare the contours
[28]:
Park19_chains = np.load(f'{data_dir}Park19/Park19_chains.npz')
print(Park19_chains['params'])
# Make posteriors from the covariance matrix
mean = fid_params_Park19.copy()
cov = Finv_PS_Park19.copy()
fisher_chain = np.random.multivariate_normal(mean, cov, size=10000)
# Corner plot
fig = plt.figure(figsize=(15,15))
colors = [col_mcmc,col_P19]
lws = [1.5,3]
# Plot 2 sigma confidence interval (https://corner.readthedocs.io/en/latest/pages/sigmas.html)
levels = 1.0 - np.exp(-0.5 * np.array([1,2,]) ** 2)
for cc, chain in enumerate([Park19_chains['chains'],fisher_chain]):
if cc == 0:
# MCMC
ls='solid'
lw=lws[cc]
hist_kwargs = {'lw':lw,'ls':ls,'density':True}
color=colors[cc]
smooth=None
fill_contours = False
no_fill_contours = True
contour_kwargs = {'linewidths':lw,'linestyles':ls}
contourf_kwargs={}
zorder=10
else:
# fisher
ls='solid'
lw=lws[cc]
hist_kwargs = {'lw':lw,'density':True,'histtype':'stepfilled', 'alpha':0.8}
fill_contours=True
no_fill_contours=False
color=colors[cc]
smooth=1
contour_kwargs = {'linewidths':0.}
contourf_kwargs = {}
zorder=0
corner.corner(chain, fig=fig,
labels=fid_labels_Park19,
smooth=smooth,
color=color, use_math_text=True,
plot_datapoints=False, plot_density=False,
no_fill_contours=no_fill_contours, fill_contours=fill_contours,
hist_kwargs=hist_kwargs,
contour_kwargs=contour_kwargs,contourf_kwargs=contourf_kwargs,
levels=levels,
range=[1,1,1,1,1,1,(40.,41.),(300,700)], # throws out a couple of outlier points in the chains [better for Lx]
show_titles=True,
zorder=zorder
);
# Format the quantile display
ax = np.reshape(fig.axes, (chain.shape[1],chain.shape[1]))
p21fish.title_double_ellipses(axes=ax, labels=fid_labels_Park19,
chain=chain,
med=None, sigma=None,
title_fontsize=18, title_pad=58,
vspace=cc/5,
color=color
)
lab_P19 = mlines.Line2D([], [], color=col_mcmc, ls='solid', lw=lws[0]+1, label='Park+19 MCMC')
lab_TW = mlines.Line2D([], [], color=col_P19, lw=lws[1]+1, label=r'This Work - Fisher Matrix')
fig.get_axes()[5].legend(handles=[lab_P19, lab_TW], loc='center left', fontsize=24)
plt.savefig(figs_dir+'corner_Park19_fisher_compare_2sigma.png', bbox_inches='tight')
['F_STAR10' 'ALPHA_STAR' 'F_ESC10' 'ALPHA_ESC' 'M_TURN' 't_STAR' 'L_X'
'E0']
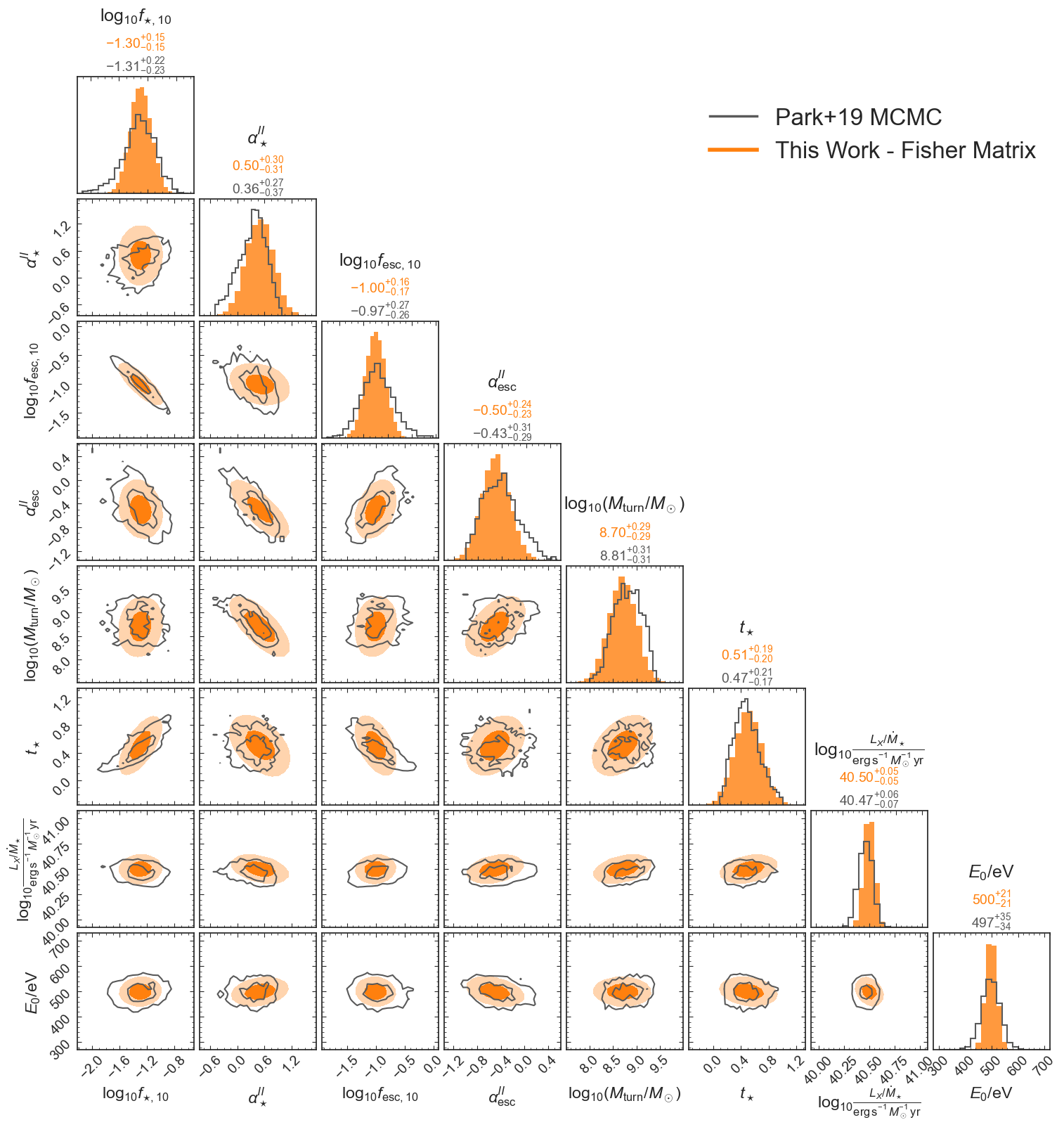
Adding a new parameter
If you want to add your own new parameter, you should:
Create lightcones varying that parameter.
Create a config file for the parameters you want to change (take one of the examples in
../21cmFAST_config_files/and replace theastro_params_varylist with your list of new parameters.Note that the fiducial parameter value for your new parameter will be the 21cmFAST default unless the fiducial value is specified in the config file. If you want a non-default fiducial parameter value you will need to create a new set of lightcones with your parameter’s fiducial included in
astro_params.Create the lightcones using
scripts/make_lightcones_for_fisher.py
Load your new parameter by adding it to the dictionary as above
See more details on running scripts/make_lightcones_for_fisher.py in the docs
[ ]:
[ ]: